–
Background
About two decades ago we described Bayesian inference for a latent class model with random effects, motivated by an application to estimate diagnostic accuracy and disease prevalence in the absence of a perfect reference test (Dendukuri and Joseph 2001). The random effects served to model conditional dependence between imperfect diagnostic tests, i.e. to model the possibility that the imperfect tests are jointly false positive or false negative. The form of the prior distributions we used at that time for the sensitivities and specificities was unnecessarily complicated, relying on a bisectional search. A more straightforward approach is described in the current article. Though the text below refers to the sensitivity, it applies equally to the specificity.
Model
The sensitivity of the ith individual is expressed as
Visualizing the distribution of sensitivity
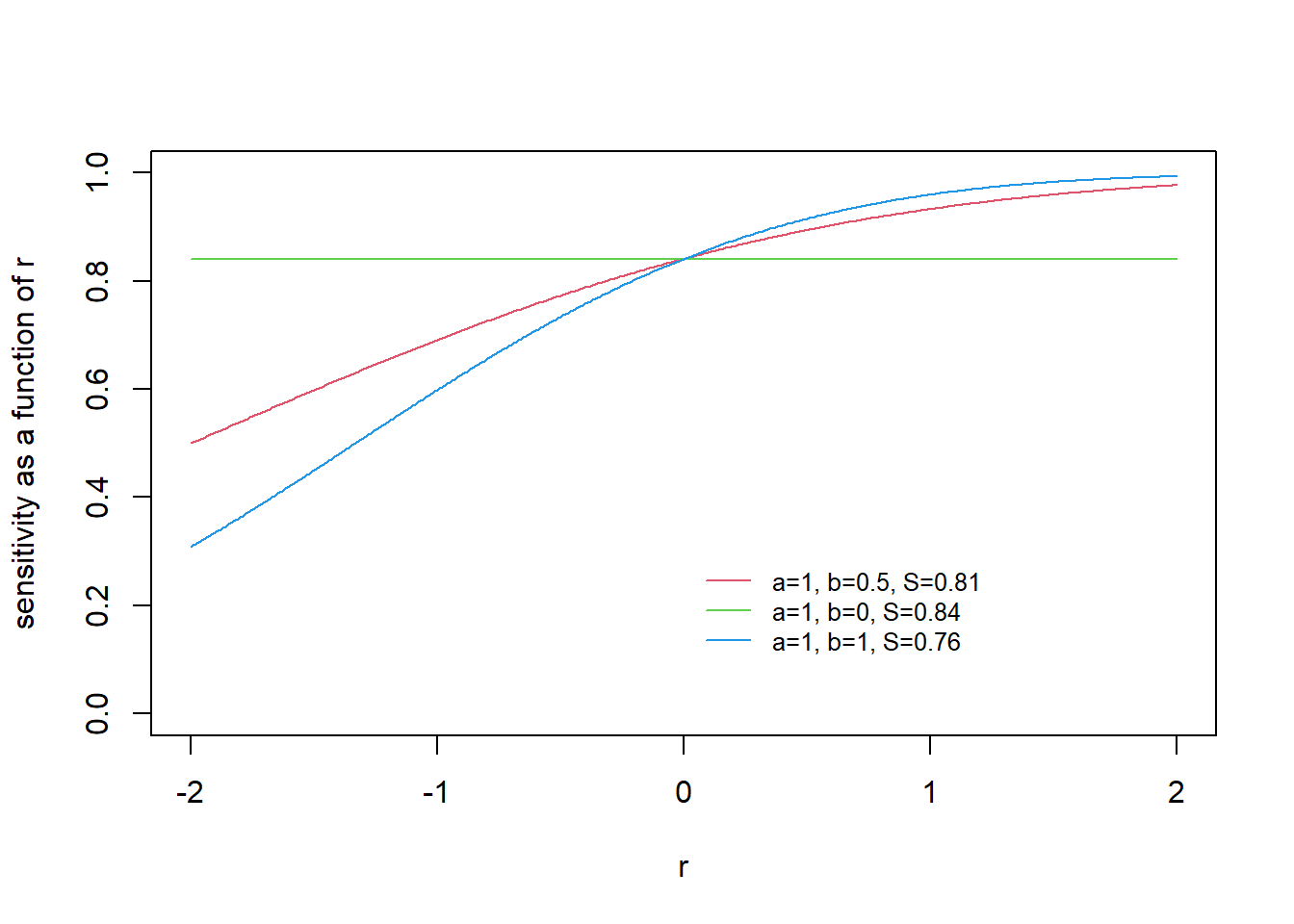
Consider the case when
In this case the marginal sensitivity
If
Prior information
Prior information is typically available on the marginal sensitivity (
Translating prior information into prior distributions
Keeping in mind that
Example
We now apply the ideas above to the Strongyloides infection example reported in the paper by Dendukuri and Joseph. The equal-tailed 95% prior credible interval over the marginal sensitivity of microscopy was (7%, 47%). In the absence of any prior information on the individual sensitivities we could assume a very wide range from (0%, 100%). This is equivalent to saying that there are some individuals in whom the test has 0% sensitivity and at the other extreme there are patients in whom it has a 100% sensitivity. It is possible that such a gradation is created by the severity of infection. Patients with a mild infection may have a very low count of the parasite that is not detectable by microscopy and those with a severe infection and correspondingly high count are always detected.
We first look at the case when
b=runif(1000,0,3)
S=rbeta(1000,4.44,13.31)
a=qnorm(S)*sqrt(1+b*b)
r=rnorm(1000,0,1)
S_i=pnorm(a+b*r)The figure below illustrates the histograms of
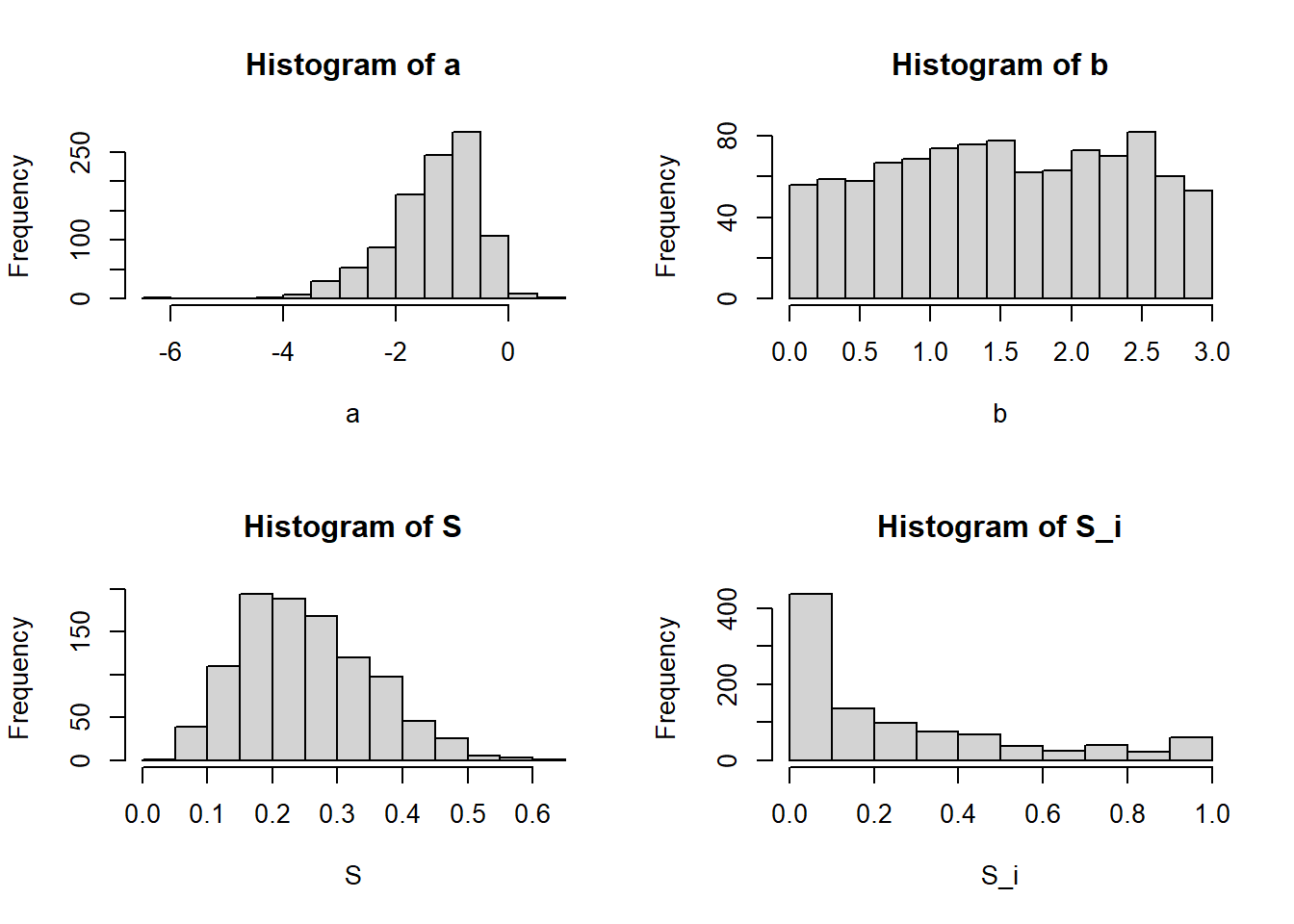
A look at the mean and quantiles of
round(quantile(S,c(0.025,0.5,0.975)),2) 2.5% 50% 97.5%
0.09 0.24 0.47 round(quantile(S_i,c(0.025,0.5,0.975)),2) 2.5% 50% 97.5%
0.00 0.15 0.99 round(mean(S),2)[1] 0.25round(mean(S_i),2)[1] 0.26The figures below illustrates the consequences of using
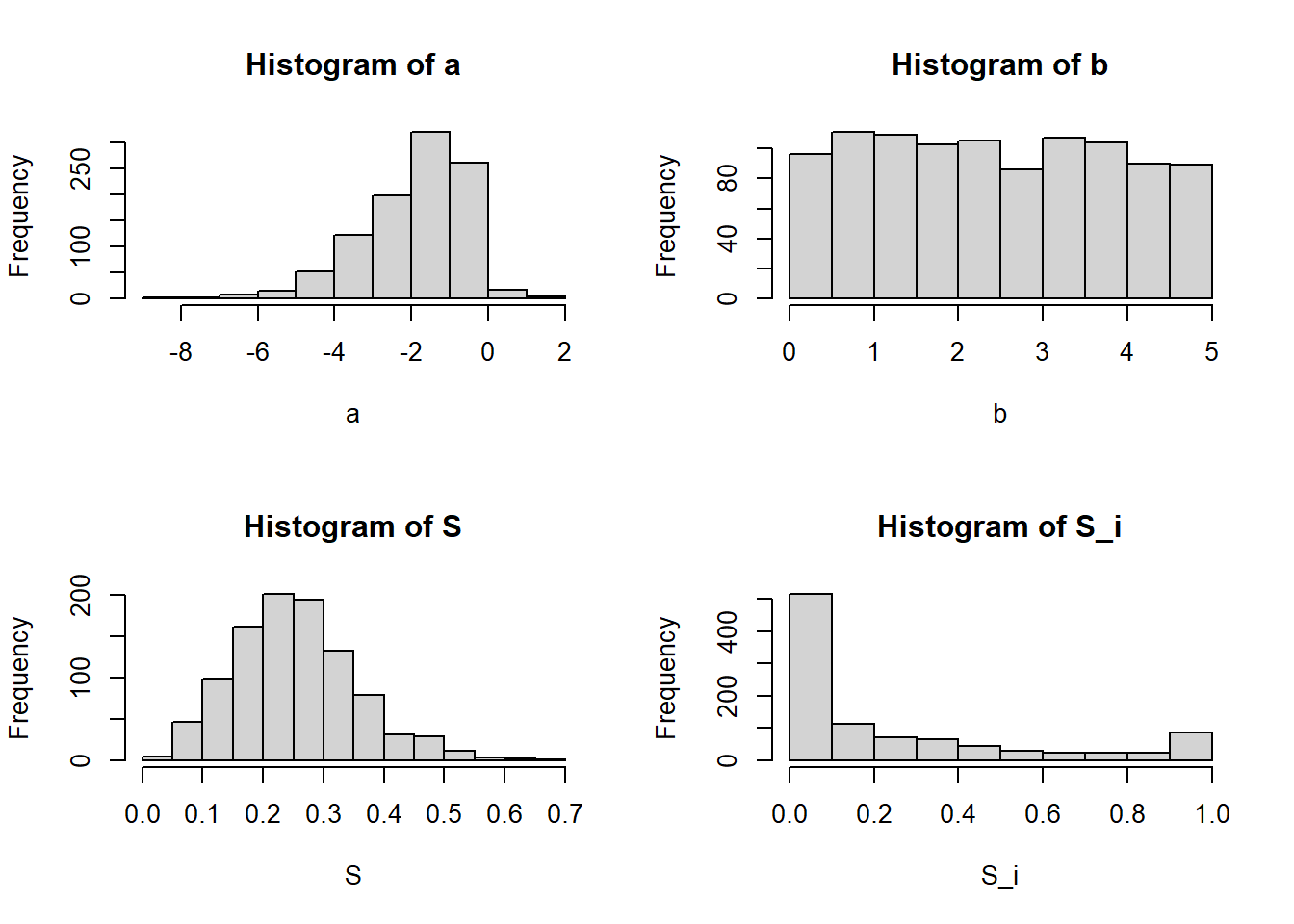
Once again, the mean values of
round(quantile(S,c(0.025,0.5,0.975)),2) 2.5% 50% 97.5%
0.08 0.25 0.48 round(quantile(S_i,c(0.025,0.5,0.975)),2) 2.5% 50% 97.5%
0.00 0.09 1.00 round(mean(S),2)[1] 0.25round(mean(S_i),2)[1] 0.24Finally, here is an illustration of the case when we have a non-informative, Uniform(0,1) prior over
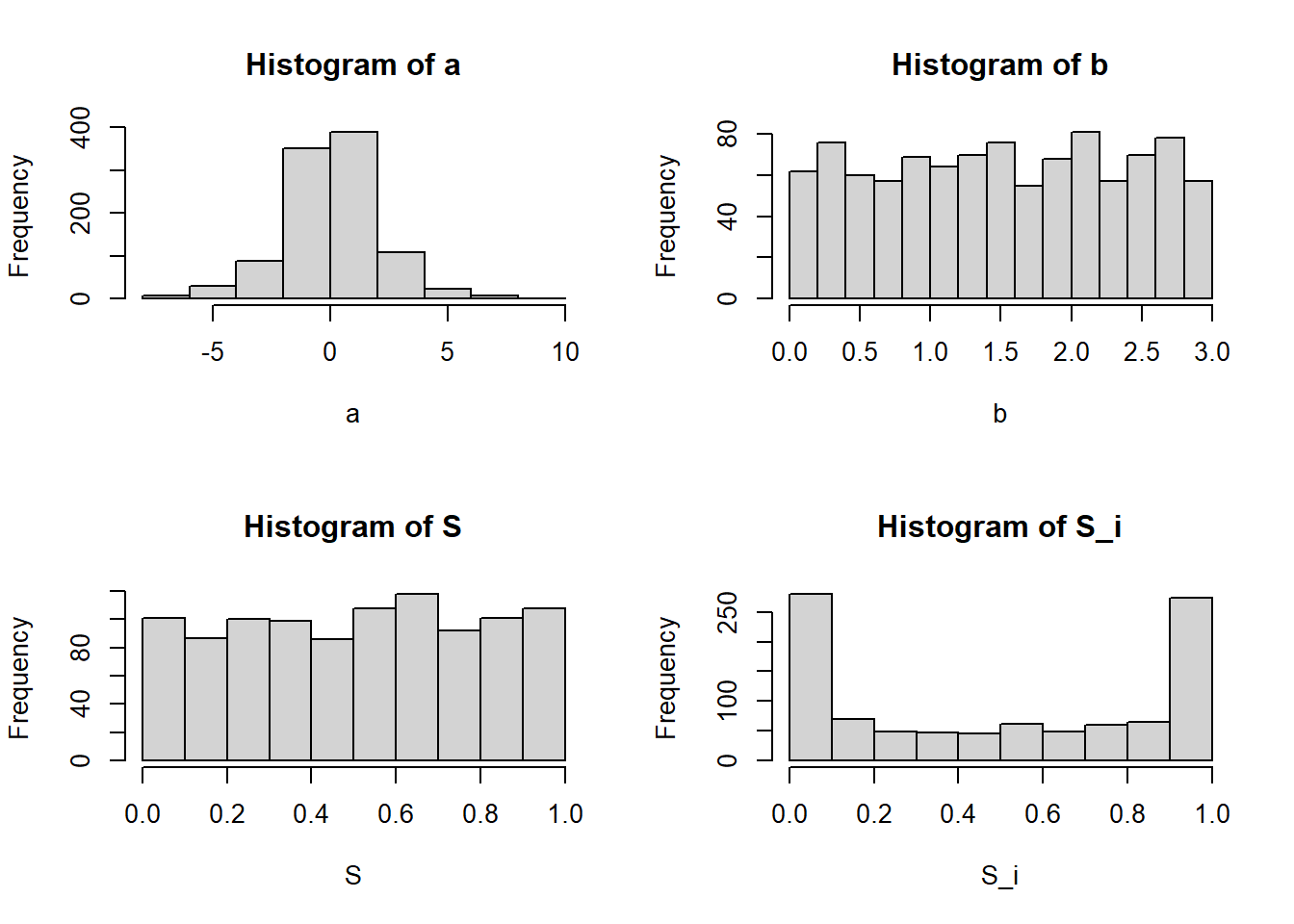
round(quantile(S,c(0.025,0.5,0.975)),2) 2.5% 50% 97.5%
0.02 0.52 0.98 round(quantile(S_i,c(0.025,0.5,0.975)),2) 2.5% 50% 97.5%
0.00 0.51 1.00 round(mean(S),2)[1] 0.51round(mean(S_i),2)[1] 0.5Summary
The approach described above is easier to implement. It has the additional advantage of ensuring the prior distribution on the marginal sensitivity
References
Citation
@online{dendukuri2022,
author = {Nandini Dendukuri},
title = {Specifying Prior Distribution for Diagnostic Test Accuracy in
Latent Class Model with Random Effects},
date = {2022-07-30},
url = {https://www.nandinidendukuri.com/blogposts/2022-07-30-lca-remodel-prior/},
langid = {en}
}